3. Enzyme Engineering
Directed evolution of the 3-hydroxypropionic acid production pathway byengineering aldehyde dehydrogenase using a synthetic selection device
Metabolic Engineering, 2018, 47, 113-120.

3-Hydroxypropionic acid (3-HP) is an important platform chemical, and biological production of 3-HP fromglycerol as a carbon source using glycerol dehydratase (GDHt) and aldehyde dehydrogenase (ALDH) has beenrevealed to be effective because it involves a relatively simple metabolic pathway and exhibits higher yield andproductivity than other biosynthetic pathways. Despite the successful attempts of 3-HP production from glycerol,the biological process suffers from problems arising from low activity and inactivation of the two enzymes.To apply the directed evolutionary approach to engineer the 3-HP production system, we constructed a syntheticselection device using a 3-HP-responsive transcription factor and developed a selection approach for screening 3-HP-producing microorganisms. The method was applied to an ALDH library, specifically aldehyde-binding sitelibrary of alpha-ketoglutaric semialdehyde dehydrogenase (KGSADH). Only two serial cultures resulted in enrichmentof strains showing increased 3-HP production, and an isolated KGSADH variant enzyme exhibited a2.79-fold higher catalytic efficiency toward its aldehyde substrate than the wild-type one. This approach willprovide the simple and efficient tool to engineer the pathway enzymes in metabolic engineering.
Engineering an aldehyde dehydrogenase toward its substrates, 3-hydroxypropanal and NAD+, for enhancing the production of 3-hydroxypropionic acid
Scientific Reports, 2017, 7, 17155
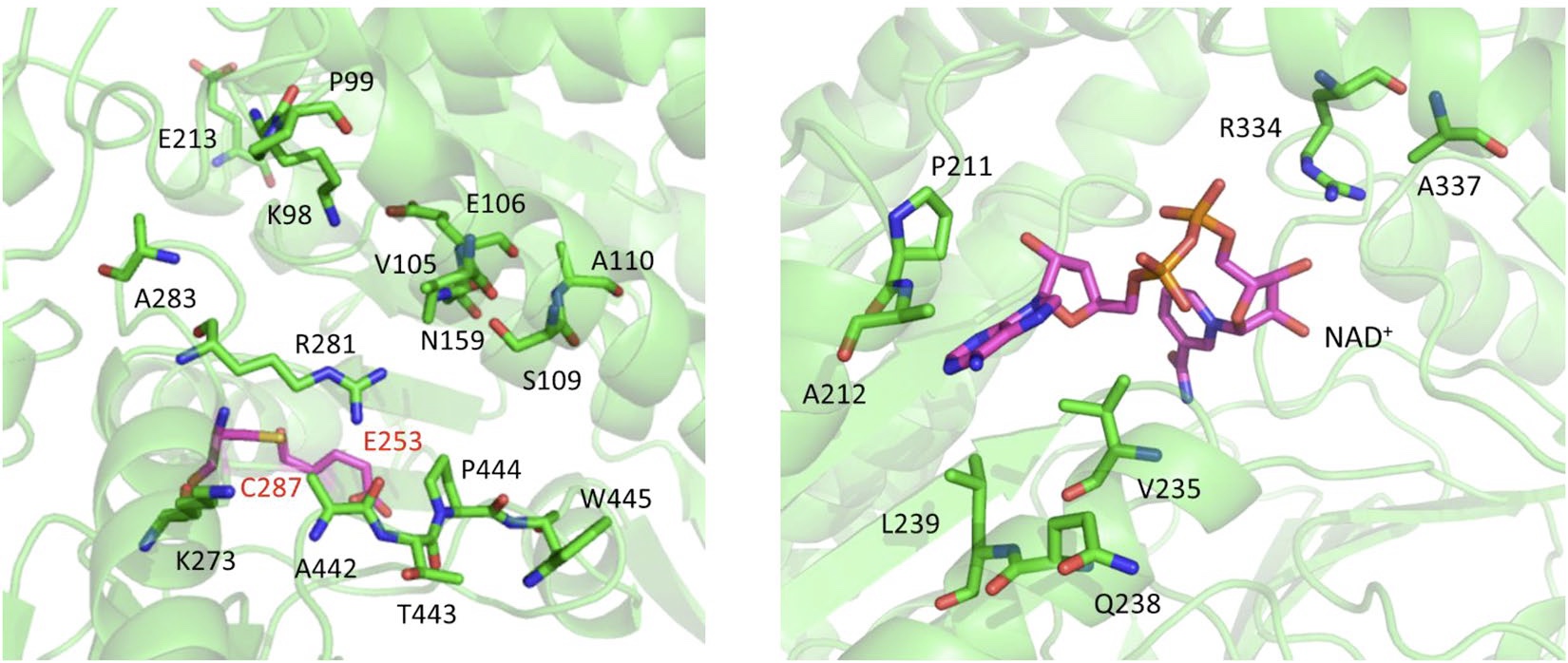
3-Hydroxypropionic acid (3-HP) is an important platform chemical, and biological production of 3-HP fromglycerol as a carbon source using glycerol dehydratase (GDHt) and aldehyde dehydrogenase (ALDH) has beenrevealed to be effective because it involves a relatively simple metabolic pathway and exhibits higher yield andproductivity than other biosynthetic pathways. Despite the successful attempts of 3-HP production from glycerol, the biological process suffers from problems arising from low activity and inactivation of the two enzymes. To apply the directed evolutionary approach to engineer the 3-HP production system, we constructed a syntheticselection device using a 3-HP-responsive transcription factor and developed a selection approach for screening 3-HP-producing microorganisms. The method was applied to an ALDH library, specifically aldehyde-binding sitelibrary of alpha-ketoglutaric semialdehyde dehydrogenase (KGSADH). Only two serial cultures resulted in enrichmentof strains showing increased 3-HP production, and an isolated KGSADH variant enzyme exhibited a2.79-fold higher catalytic efficiency toward its aldehyde substrate than the wild-type one. This approach will provide the simple and efficient tool to engineer the pathway enzymes in metabolic engineering.

Synthetic Protein Engineering Laboratory
Department of Molecular Science and Technology, Ajou University
206 Worldcup-ro, Yeongtong-gu, Suwon, 16499 Korea